Overview
The goal of the Cowley Lab is to understand how the developmental environment impacts the epigenetic regulation of the genome to program metabolic disease susceptibility in later life.
Epigenetic mechanisms have been widely proposed as mediators of disease programming. Our lab proposes that epigenetic dysregulation of imprinted genes plays an essential role in connecting the developmental environment to later life disease for three reasons:
To understand how imprinted genes link the developmental environment to metabolic health, we study two important developmental stressors: maternal metabolic syndrome (MetS) and cadmium (Cd).
MetS is defined as a cluster of co-occurring conditions including obesity, high blood pressure and high circulating triglyceride levels, that increase the risk of heart disease, stroke and type II diabetes. MetS affects 30 % of US adults and is of major concern for the health of the next generation given that 26 % of pregnancies begin with a body mass index >30.
The heavy metal Cd is one of the top ten toxicants of major public health concern identified by the World Health Organization. The primary sources of exposure are cigarette smoking and the consumption of contaminated crops or water.
Epigenetic mechanisms have been widely proposed as mediators of disease programming. Our lab proposes that epigenetic dysregulation of imprinted genes plays an essential role in connecting the developmental environment to later life disease for three reasons:
- We and others have shown that the unique epigenetic mechanisms at Imprinting Control Regions (ICRs), which regulate the monoallelic expression of imprinted genes, are particularly susceptible to perturbation by the environment in early life;
- Once established, the epigenetic profiles at ICRs persist throughout life providing a long-term memory of developmental exposures;
- Imprinted genes play critical roles in regulating metabolism in adulthood.
To understand how imprinted genes link the developmental environment to metabolic health, we study two important developmental stressors: maternal metabolic syndrome (MetS) and cadmium (Cd).
MetS is defined as a cluster of co-occurring conditions including obesity, high blood pressure and high circulating triglyceride levels, that increase the risk of heart disease, stroke and type II diabetes. MetS affects 30 % of US adults and is of major concern for the health of the next generation given that 26 % of pregnancies begin with a body mass index >30.
The heavy metal Cd is one of the top ten toxicants of major public health concern identified by the World Health Organization. The primary sources of exposure are cigarette smoking and the consumption of contaminated crops or water.
Current projects
Imprinted genes in the programming of non-alcoholic fatty liver disease (NAFLD) by maternal MetS
NAFLD in children is a major public health concern, affecting 3-10 % of the pediatric population. Within the next decade, juvenile NAFLD is predicted to become the most prevalent cause of liver failure in childhood in developed countries.
|
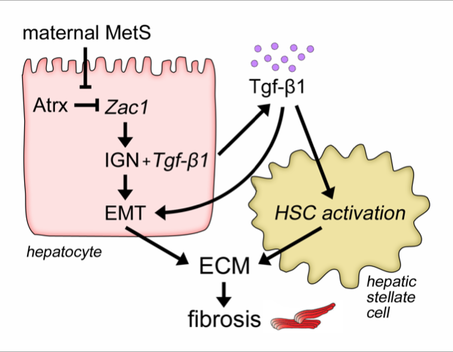
Having a mother with MetS during perinatal life is a major risk factor for juvenile NAFLD. However, the underlying mechanisms and critical periods of susceptibility during development remain unclear.
Using a combination of diverse in vivo and in vitro systems, we have identified an Imprinted Gene Network (IGN) driven by the transcription factor Zac1 that programs susceptibility to juvenile NAFLD in response to maternal MetS. Our ongoing work aims to identify the cellular mechanisms through which activation of these genes drives NAFLD, and to determine the epigenetic basis for their dysregulation by maternal MetS.
Using a combination of diverse in vivo and in vitro systems, we have identified an Imprinted Gene Network (IGN) driven by the transcription factor Zac1 that programs susceptibility to juvenile NAFLD in response to maternal MetS. Our ongoing work aims to identify the cellular mechanisms through which activation of these genes drives NAFLD, and to determine the epigenetic basis for their dysregulation by maternal MetS.
|
Related publications:
|
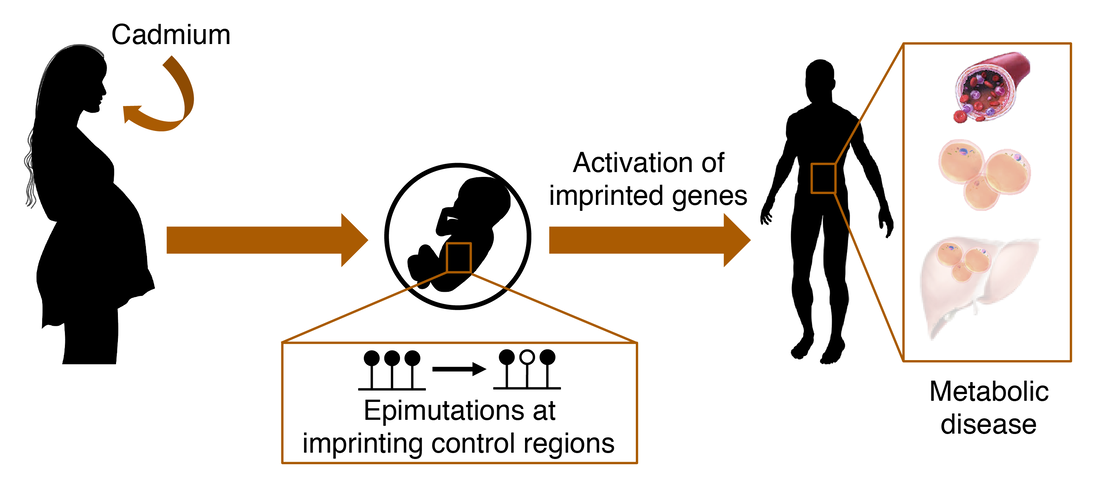
Epigenetic mechanisms linking developmental Cd exposure to disease in adulthood
In addition to the widely-studied health effects of exposure to Cd during adulthood, we have found that exposure to Cd during development causes behavioral abnormalities, including impaired social behavior, and programs increased susceptibility to metabolic disease, including NAFLD and elevated blood pressure.
|
Using an integrated approach of unbiased high-throughput screens (transcriptomics, methylomics, proteomics, metabolomics) and targeted assays, we are determining the underlying epigenetic and molecular mechanisms that link developmental Cd exposure to adult disease. We hypothesize that imprinted genes play a central role in this process.
|
Related publications:
|
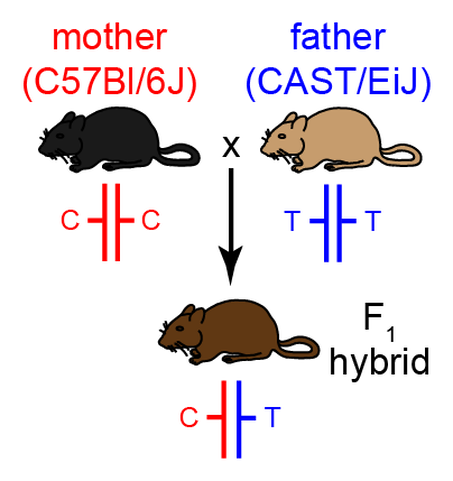
Using a unique genetic mouse model, we are studying how Cd affects imprinted gene regulation in the placenta, and are investigating novel epigenetic, transcriptional and molecular mechanisms that mediate Cd-induced placental dysfunction. Specifically, our model allows the parental alleles of imprinted genes to be distinguished and studied independently, providing novel insight into the mechanisms of Cd action.